For years scientists built on Sanger's chain-termination method to develop new technologies. Over time more efficient techniques evolved, thus blossomed the next generation of DNA sequencing. Today there are quite a few individual methods, and still more are developing. The leading company's are competing for the fastest and most efficient sequencing technologies. The competition promotes rapid advances and breakthroughs in new methods.
Click on a company's logo for the link to their website.
Massively Parallel Signature Sequencing (MPSS)
The first of the "next-generation" sequencing technologies, MPSS, was developed in the 1990's by Lynx Therapeutics, a company founded by Sydney Brenner and Sam Eletr. MPSS uses a bead-based method and involves complex adapter ligations followed by adapter decoding, reading the sequence in increments of four nucleotides. This method was complicated and susceptible to sequence-specific bias, so no machines were sold. Lynx Therapeutics only preformed "in house" sequences. After the development of sequencing-by-synthesis, MPSS became obsolete. (5)
Pyrosequencing
| A parallelized version of pyrosequencing was developed by 454 Life Sciences, and since then it has been acquired by Roche Diagnostics. This next generation sequencer technology was the first to achieve commercial introduction, in 2004. In pyrosequencing, each addition of a nucleotide by DNA polymerase releases a pyrophosphate. This initiates a series of reactions that ultimately produce light by the enzyme luciferase. In the Roche/454 method, fragments of DNA are amplified using the surface of agarose beads. These amplified single molecules are then sequenced by filing into a picotiter plate, each bead in one of the several hundred thousand wells, and the 454 base-calling software is able to calibrate the light emitted by a single nucleotide incorporation. (6) |
Illumina Sequencing
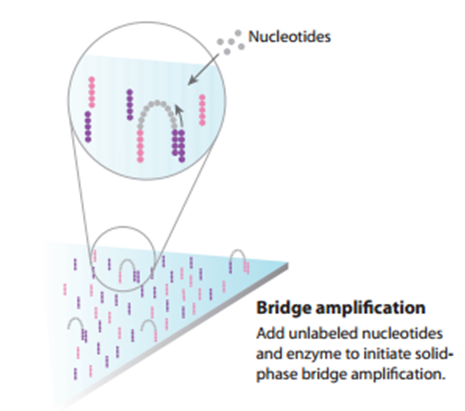
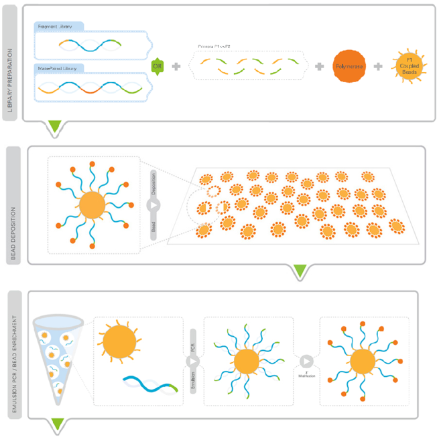
A leader in the market for sequencing technologies is the company Illumina. Illumina uses a sequencing process inspired by the concept of capillary electrophoresis, however its next generation sequencing technology enables rapid sequencing of long stretches of DNA base pairs spanning entire genomes. Their latest technologies are capable of producing hundreds of gigabases of data in a single sequencing run. To achieve such huge amounts of data, the process starts with the use of bridge amplification to create a library of small DNA segments. (6) The segments then can be uniformly and accurately sequenced by identifying signals emitted from fluorescently-labeled base pairs. The newly identified strings of bases, called reads, are then reassembled using a known reference genome as a scaffold. The full set of aligned reads produces the entire sequence of each chromosome in the DNA sample. (7)
| Figure (4) |
Applied Biosystems SOLiD sequencer
Applied Biosystems SOLiD technology employs sequencing by ligation. Here, a pool of all possible oligonucleotides of a fixed length are labeled according to their sequence position. (15) Oligonucleotides are annealed and ligated and the preferred ligation by DNA ligase for matching sequences indicates which nucleotide is at that position. Before sequencing, the DNA is amplified by emulsion PCR. The resulting bead, each containing only copies of the same DNA molecule, is deposited on a glass slide. The result is sequence of large quantities of DNA, making it a very efficient form of sequencing.(5)
Ion Semiconductor Sequencing
Ion Torrent, owned by Life technologies, created a line of sequencing machines that are extremely fast at sequencing, yet small enough to be placed on a desk. The system is based on using standard sequencing chemistry (PCR), but with a novel, semiconductor based detection system. The system is based upon detection of hydrogen ions that are produced during DNA replication, as opposed to the visual methods used in other sequencing methods. A template DNA strand is placed in a microwell which is then flooded with a single type of nucleotide. If the introduced nucleotide is complementary to the leading template nucleotide, it is incorporated into the growing complementary strand. This causes a reaction that releases a hydrogen ion which triggers a hypersensitive ion sensor. If homopolymer repeats are present in the template sequence, multiple nucleotides will be incorporated in a single cycle. This results in a corresponding number of hydrogens and a proportionally higher electronic signal. The two machines by Ion Torrent, the Ion Proton Sequencer and the Ion Personal Genome Machine, are both powered by semiconductor chip technology. (27)
Ion Proton Sequencer Figure Ion Personal Genome MachineFigure (19) | 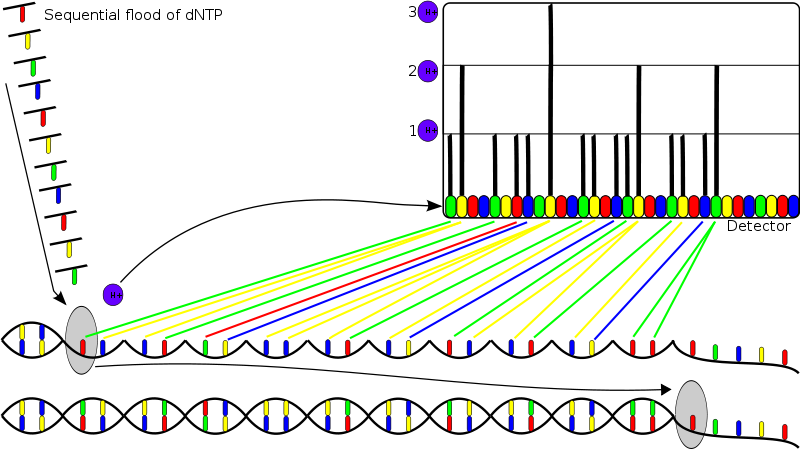 Released hydrogen ions are detected by an ion sensor. Multiple incorporations lead to a corresponding number of released hydrogens and intensity of signal. Figure (28) Semiconductor Sequencing Chip (29) |
Sequencing with Nanopores
Nanopores are simply holes with a diameter of one nanometer. When a nanopore is immersed in a fluid and a voltage is applied ions are conducted through the nanopore causing an electrical current. The ability to observe this electrical current forms the basis of DNA nanopore sequencing. The method starts with DNA passing through the nanopore, either by electrophoresis or an enzyme guiding the strand through the pore. As the DNA passes through the hole, the bases interrupt the electrical signal. The amount of current which can pass through varies depending on which base is currently passing through the nanopore. (30) Recently, Oxford Nanopore Technologies has utilized this technology to create two new sequencing machines to come out this year, 2012. The GridION, and the more revolutionary MinION. (32) The MinION is a tiny, disposable USB-key sequencer that can sequence about a billion base pairs of DNA, and cost around $500-$900 each. A sequencer this accessible would transform the field of biology. This sort of technology is referred to as the third-generation sequencing technology. (31)